Go to first part: http://plin9k.ucoz.com
Table 2. Transcripts
with traces of an EcoRI related linker
and some related references.
# |
Accession/Organism |
Gene/Protein * |
Linker ** |
References *** |
|
1 |
U58090 Homo sapiens |
Cullin gene family member, Hs-cul-4A |
aattcggcacgagctcgtgccgct NSARARAA |
Cell
85, 829-839. 1996. |
|
2 |
U28831 Homo sapiens |
Protein immuno-reactive with anti-PTH polyclonal antibodies |
gcacgagctcgtgccgat ARARAD |
Proc. Assoc. Am. Physicians 107, 296-305. 1995. |
|
3 |
BC041619 Homo sapiens |
Protein KIAA0404, for IMAGE:5923662 [R: hypoth. prot. MGC16044] |
ccctcgtgccgaattcggcacgag PSCRIRHE |
Proc. Natl. Acad. Sci. U.S.A. 99, 16899-16903. 2002. |
|
4 |
AF176705 Homo sapiens |
F-box protein FBX10 (PINX1) [R: vector] |
cctcgtgccgaattcPRAEF |
Curr. Biol. 9, 1180-2. 1999. |
5 |
X85792 Homo sapiens |
Vpr binding protein 1 |
tcgtgccgaattcggcacgag SCRIRHE |
Benichou et al. [Unpublished]; J Biol Chem. 277, 45091-8. 2002 |
6 |
AF151109 Homo sapiens |
Putative BRCA1-interacting protein (BRIP1) |
ggcacgagctcgtgccgc GTSSCR |
Wang et al. BRCA1-interacting protein. [Unpublished]; Oncogene 19, 6152-8. 2000. |
|
7 |
AF146697 Homo sapiens |
FOXP1 |
aagaattcggcacgagct KNSARA |
Cancer Res. 61, 8820-8829. 2001. |
|
8 |
NM_002342 Homo sapiens |
Gene and 3' UTR for TNFR
superfamily, member 3 (LTBR) |
gctcgtgccgaattc |
Genomics 16, 214-218. 1993. [Curated by NCBI] |
|
9 |
X93499 Homo sapiens |
RAB7 protein,
GTP-binding [L: Dystroglycan 1. C:
Rab7. R: Envelope glycoprotein] |
ccccgaattcgggg & gcccgaattcgggc |
Biochem. Biophys. Res. Commun. 229, 887-890. 1996. |
|
10 |
X82200 Homo sapiens |
Gene and mRNA for interferon-induced Staf50 |
gaattcggcacgagctc |
J. Biol. Chem. 270, 14891-14898. 1995. |
|
11 |
U31384 Homo sapiens |
mRNA for G protein gamma-11 subunit |
ggcacgagctcgtgccg |
J. Biol. Chem. 270, 21765-21771. 1995. |
|
12 |
AF379619 Homo sapiens |
Intron near AB13, precursor mRNA |
gaattcggcacgagct |
van Roy and Staes. New
human gene family. [Unpublished]. |
|
13 |
AY245868 Homo sapiens |
CDS for Aldehyde
oxidase-like protein (AOX2) pseudogene |
aagaattcggcacgagca LNSARA |
Wright RM. Human aldehyde
oxidase. [Unpublished]. |
|
14 |
AF339764 Homo sapiens |
mRNA from Fetal liver spleen IMAGE:108721 |
gaattcggcacgagcggcacgagct
|
Genomics 79, 635-656. 2002. |
|
15 |
U43527 Homo sapiens |
5'UTR for Malignant
melanoma metastasis-suppressor (KiSS-1) |
(ctct)15cctcgtgccgaattcggcacgag |
J. Natl. Cancer Inst. 88, 1731-1737. 1996; Genomics 54, 145-148. 1998. |
Notes: This table presents the EcoRI related linker ctcgtgccgaattcggcacgag as it appears in the Genbank for some human genes. To view the rest of this Table 2 and the presence of the linker in other organisms, refer to URL:
* Gene/Protein (Gene symbol) [notes for sides of linker (L or
R)]
** Linker and its Translation in Amino Acids as presented in the Genbank
*** Corresponding References According to the Genbank; closest related match
Abundance of sequences including the EcoRI-like palindromic linker
There
are thousands of sequences, including expressed
sequence tags (est) in the Genbank and in other nucleotide
databases that still contain artifacts, having as its common denominators, EcoRI palindromic linkers like the ones
described in this article.
The
palindromic linkers can be present in tandems, halves, or in different lengths;
being 12 to 24 bases its most common range. Artificial linkers have been found
even inside multiple coding regions, like the examples presented in the full
Table 2 (7). Examples of those linkers are frequently present outside the
coding region, i.e, in promoters,
like in:
1-) NR_001557 for H.
sapiens aldehyde oxidase 2 (AOH2) on chromosome 2, oligo gaattcggcacgagc
(13).
2-) NM_002342 H. sapiens lymphotoxin beta-receptor (LTBR; member 3 of the TNFR superfamily), oligo gctcgtgccgaattc (14).
Furthermore, those palindromic linkers have been found also in the 5' region, i.e., in sequence U43527 for the human malignant melanoma metastasis-suppressor KiSS-1, oligo (ctct)15cctcgtgccgaattcggcacgag (15), and/or in the 3' region, i.e., sequence AY029161 for the Pin2-interacting protein X1, oligo ctcgtgccgaattcggcac (16).
Of the
few submissions to the Genbank that
are explicitly reporting the presence of the EcoRI adapter, Hirama et al
(17) stands out, together with Inoue et
al (18) and Savas et al (19).
However, Inoue et al (18) considers
only the 8 first bases at the left flank as part of the sequence for the EcoRI-adapter. The effect of the
palindromic linker for Inoue's sequence may extend to at lest 16 bases
(sequence D83948 for S1-1 protein, oligo ggcacgagctcgtgccg) by an apparent
phenomenon of self-recombination and self-insertion inside the host-vector
interactions.
A similar
situation to what we see in Inoue's is presented by Savas et al (19), mentioning in their Genbank
submission the first 6 bases only as part of the EcoRI-adapter (same in reference 1, but not in the submitted
sequence L21934). In Savas' reference (19) the linker-like effect may be
extended about 20 bases (sequence X78445 for Cytochrome P450 Cyp1-b-1, oligo gaattcggcacgaactcgtgc).
Hirama et al (17) is the only one
that appropriately mentions a longer extension for the EcoRI-adapter, reporting it as being of 14 bases (sequence X56703
for the rearranged T-cell receptor alpha chain, oligo gaattcggcacgagct).
Chimaeras linked by the EcoRI-like palindrome seem to be resistant to enzymatic digestion
The
palindromic linkers persist in the sequences without being digested by the
enzymes. The discovery of mechanisms of resistance to the enzymatic digestion
awaits further study. However, it is evident that the most common palindromic
linkers match the identity of the EcoRI
adapter sequence gaattcggcacgag, which is used for the 5'UTR, and reported to
the Genbank, i.e., inside the sequence AI607511 as ctcgtgccgaattcggcacgag (similar
to acidic ribosomal phosphoprotein PO). In that sequence, it is indicated the
use of the vector pBluescript SK(-),
plus EcoRI, with the additional use
of the vector Uni-ZAP XR. These
methodologies, like the ones described in (1, 5, 6), may be promoting the
phenomenon of artificial linkage abundantly present in the Genbank.
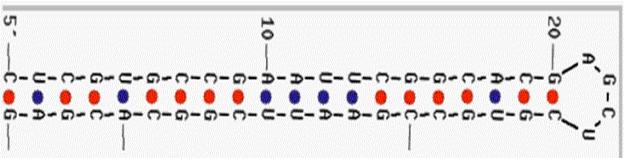
The rearrangement and splicing in a host-vector interaction resistant to EcoRI enzymatic digestion may be explained by a phenomenon of self-hybridization performed by the linker (Figure 3), which could make it to appear as an appendage impossible to be grasped by the digestive enzyme.
Figure 3.
Self-hybridization of the palindromic EcoRI-like
linker seems to block enzymatic digestion. Phenomenon also seen as 'a closing zipper', at both ends of longer
sequences, where two distant parts of the linker approach and stick together
producing a plasmid-like formation. [Examples 38-42, full Table 2 (7). Figure
obtained using the software from reference 20.]
Go to part 3: http://plin9k.ucoz.com/index/0-27




