Palindromati
This work in a better quality format and with more space can be found at: http://www.reocities.com/plin9k
Fernando Castro-Chavez.
fdocc at yahoo dot com
Independent Biotechnologist.
ABSTRACT
This article describes a family of artificial
heterotranscripts (RNA chimaeras) composed by thousands of Genbank sequences containing fragments or the complete EcoRI-like adapter acting as the
palindrome linker ctcgtgccgaattcggcacgag,
binding together two or more genes that may be produced by different
chromosomes. This happens due to current methodologies producing the reported
sequences, found in the Genbank, in Affymetrix microarrays, and in many
published articles reporting or using those sequences that include the EcoRI-like linker inside coding regions,
and/or 5'UTR or 3'UTRs mRNA sites. This EcoRI-like linker and its heterotranscripts are
here deemed as experimental artifacts, characterization that can be helpful to
prevent errors, both in the studies of molecular mechanisms and in the drug
discovery process.
Key words: EcoRI, Palindrome,
Perilipin, Genbank, Affymetrix, microarray, species-specific.
INTRODUCTION
It is
vital in the discovery of new medical treatments to target precise molecules
without having side effects for organic tissues. To accomplish this objective
it is necessary a stringent quality control within molecular databases. This
article describes the finding of numerous methodological artifacts reported to
the Genbank. It is recommended a most
carefully analysis of nucleic acid sequences for biological, medical and drug
discovery purposes.
A single RNA binding in one-strand two different genes from two different human chromosomes (1) was the theoretical beginning for the study on heterotranscripts.
Here, I
define heterotranscripts as
chimaeras, sequences composed by fragments corresponding to two or more genes
from the same or from different chromosomes.
I thought that such a phenomenon reported in reference (1) must have been reflected in a rational and logical combination of Intelligently Designed gene products (2, 3). As most of the vital molecules and biological pathways are present in many organisms, I initially thought that the phenomenon described in reference (1) maybe should be present in many natural sequences as a possible functional common denominator.
I initially supposed that the study of sequences similar or related to the one present in reference (1) maybe could be helpful for our understanding of the molecular basis to biological change.
Thus, this particular phenomenon was a possible prospect for the abundance of proteins exceeding their number of genes via multiple modular combinations of diverse mRNAs. Recent estimates for humans reckon above a million of proteins produced by just 20,000 to 25,000 genes (4).
With these considerations in mind, my initial idea was that, if reference (1) was true, then the production of those numerous proteins could have had a putative process of RNA hetero-linkage at their formation.
However, after five years of comparing sequences, I came to realize that those hetero-sequences using that same oligonucleotide as its common linker were just methodological artifacts.
The
common element of these chimeras is the linker ccgaattcgg (as presented in reference 1 inside the
sequence L21934 for the H. sapiens
ACAT-1 enzyme). This leaves references 1, 5 and 6 (if real), as a one and
unique possible species-specific
phenomenon in humans (2, 3).
Another
article on the same sequence (1), has recently been published by the same group
(5). Its authors mentioned since reference (1) the similarity of that linker
with the EcoRI-adapter (a tool
extensively used in molecular biology research), so the door still is open to
verify whether this is a methodological artifact or not (5).
The
initial construction of that sequence demonstrates that their cDNA library was
transformed in E. coli (strain MC1061) using the phagemid vector pBluescript as well as with the
expression vector pcDNA. Then, they retransformed
it in the same E. coli strain (6), again.
However, I have recently seen that the use of similar vectors can be
involved in the production of chimerical artifacts in multiple instances, like
in those examples presented in Tables 1 and 2 (7).
A
possible, however remote, explanation for reference (1) is that we are dealing
with a natural process, mostly restricted to humans. Yet, whatever the final
verdict may be, the fact is that the EcoRI-like
linker or adapter described in (1) was the starting point for the next
findings, described in this article.
My
personal hypothesis is that heterotranscripts or chimeras including the EcoRI-related palindromic linker ctcgtgccgaattcggcacgag or its related sequences, extending
themselves to at least twelve bases, are artifacts from the molecular
methodologies used, mainly mediated by its host-vector interactions.
RESULTS AND DISCUSSION
The finding of a related palindrome in
Affymetrix microarrays
The
basis for this article appeared while working with antiobesity microarrays. By
studying the changes of gene expression in the obesity resistant perilipin
knock out mice (8, 11), with the DNA-Chip Affymetrix
MG-U74A-v2, analyzed using the free educational software dChip V.1.2 (9). One particularly intriguing hetero-transcript was
the nucleotide sequence AB030505, initially reported by its submitters as the Mus musculus mRNA for UBE-1c1, UBE-1c2
& UBE-1c3 (complete cds). The following paragraph describes the sequence
AB030505 and the common EcoRI-like
linking element present in thousands of other Genbank sequences.
A careful study of the nucleotide sequence AB030505 using Blast (10) led me to an element that was linking two large sections from two different genes:
1. The nucleotide sequence AK078792 from chromosome 10,
coding for a melanoma ubiquitous mutated protein homologue (Mum1) and
2. The nucleotide sequence BC036273 from chromosome 12, coding for retinol dehydrogenase 11 (similar to Arsdr1). The linking element within the sequence AB030505 corresponded to the palindrome ctcgtgccgaattcggcacgag, composed by 22 nucleotides.
Here again, as in the initial report (1), two transcripts originated in two different chromosomes were linked together in one mRNA strand. Those 22 bases contain the core palindromic linker ccgaattcgg at its center, which is similar to the one initially reported by reference (1).
A
palindrome sequence for the double helix of DNA has the same nucleotides if
read from 5' to 3', which is the normal reading direction, either from the plus
(+) or from the minus (-) strand. A manual and visual assessment of this palindromic
linker was done. Amazingly, this linker was present in thousands of sequences
reported to the Genbank.
In the
full Table 2 (7), I present many examples of the palindrome (or related
sequences) being reported as if they were present inside coding regions. The
palindromic linker mentioned is frequently translated as the artificial peptide
RAEFGT, absent in sequenced protein
databases (10).
Increase in the number of palindromic sequences reported to
the Genbank
A monthly increase was seen in
the number of sequences containing the EcoRI-like
linker or its derivatives inside thousands of sequences. In one recent example
(14 Oct. 2005) done in Blastn (nucleotide to nucleotide alignments),
selecting the non redundant (nr) nucleic acid database sequences of Genbank, a query of 44 palindrome
letters was used:
CTCGTGCCGAATTCGGCACGAGCTCGTGCCGAATTCGGCACGAG
With
this query, I obtainined 6010 Blast Hits using the next query conditions:
1. 106 as the minimum expected number. Some
results are presented in Table 2 (7).
2.
1000 as the number of descriptions and of alignments.
In the Genbank's alternate database containing expressed sequence tags (est),
which are mRNAs for putative proteins, the number of sequences containing the
palindromic EcoRI-adapter is also
present by the thousands.
Additional palindromes found by using microarrays
Additional targets pertaining to
these linkers were also found while studying the results of microarrays
available online using the software
tool dChip (9) coupled to the Affymetrix probes databases. Table 1
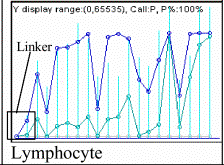
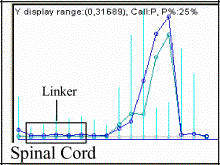
shows examples containing the palindromic linkers.
Affymetrix has been a
successful microarray methodology, i.e.,
to evaluate the gene expression in humans, mice, and rats. However, both the
presence of artificial heterotranscripts and/or of their own artificial linkers
can lead to a misrepresentation of its real expression inside the tissues, as
the area under the curve is reduced for those genetic sequences.
Table 1. The EcoRI-related palindromic linker is
present both in the Genbank sequence
targets and in Affymetrix microarray probes for humans,
mice and rats.
|
ID/
Organism |
EcoRI Affymetrix Probes from Genbank [DNA Chip] |
Graphic,
non-expression of EcoRI-linker |
Reference |
|
AB002533_at Homo sapiens |
gaattcggcacgagcacgcgtgaga, ggcacgagcacgcgtgagacttctc [in the Human DNA Chips HuGene-FL and in Hu6800] |
Ribosomal protein LP2 (Qip1)
|
Shipp et al, Nature Med. 8, 68. 2002 |
|
AJ243503 (99534_at) Mus musculus |
ttcggcacgagctcgtgccggtcct [in the Mouse DNA Chips MG-U74Av2 and in MG-U74A] |
Adipocyte Ghrelin
|
Castro-Chavez et al. Diabetes 52, 2666. 2003 |
|
AI045710 (rc_AI045710_at) Rattus norvegicus |
atgatatgtacagatccctcgtgcc, tgatatgtacagatccctcgtgccg, tatgtacagatccctcgtgccgcct, tgtacagatccctcgtgccgcctcg, gtacagatccctcgtgccgcctcgt [in the Rat DNA Chips RG-U34B} |
Disulfide Isomerase related prot. (Erp70)
|
Children's National Medical Center Accesed Feb. 1, 2005. |
Note: The EcoRI-related palindromic linker ctcgtgccgaattcggcacgag causes the
drop of microarray expression to zero demonstrating its absence in the tissues
[dChip V.1.2 (9)]. Highlighted in the
second column in clear blue are the portions corresponding to the palindromic
linker, and in dark blue, the nucleotides exchanged to obtain the second set or
"mismatch" in Affymetrix' probes (DNA-Chip).
The phenomenon of heterotranscription
Twelve
bases seem to be the minimum common denominator in order for the EcoRI palindromic linker to produce
artificial heterotranscripts such as the ones reported here and present in the
Genbank.
The most common palindromic flanks for the oligonucleotide ccgaattcgg are g and c, which give us the longer oligonucleotide gccgaattcggc. Less frequent are the flanks c and g to produce the second oligonucleotide cccgaattcggg, with a similar effect on heterotranscription. This last palindromic sequence is the one that we have in reference (1). The same palindromic sequence is present in example 9 from Table 2 (Homo sapiens X93499 for the RAB7 protein), in which we have fragments for more than two genes attached together in the same strand, through the palindromic linkers ccccgaattcgggg and gcccgaattcgggc (12).
To go to part 2: http://plin9k.ucoz.com/index/palindromati_part_2/0-26